So I have a large number of 12/12 yDNA matches, 68 to be precise.
Let’s look at what a few different places say about the significance of these matches.
FamilyTreeDNA has the TiP Report calculator.
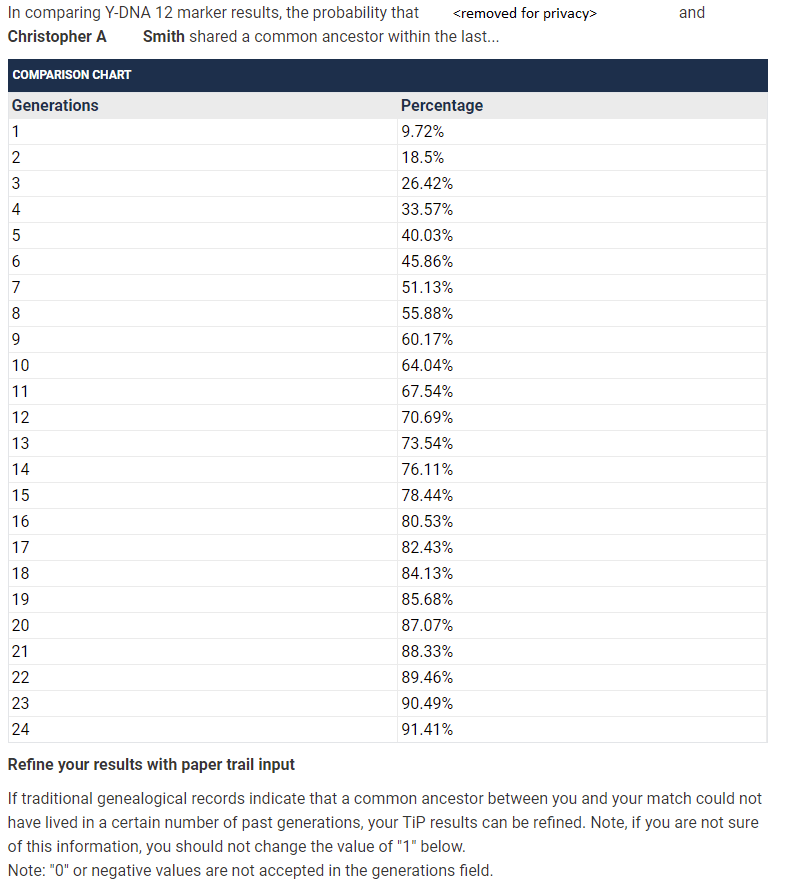
Interestingly, despite the following posting on FTDNA’s official FAQ pages, I’m still given the option to adjust the number of generations for 12/12 matches:

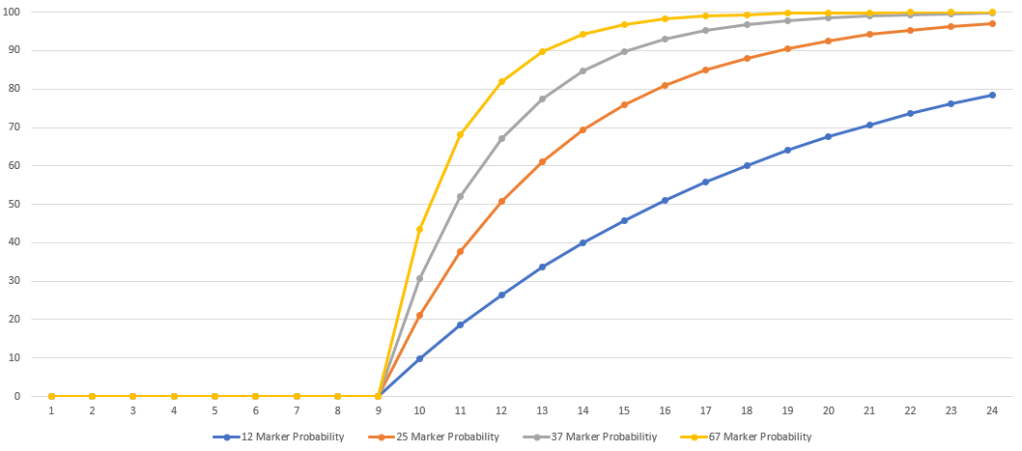
Using the default values, this curve appears like this: I’ve added 25, 37, and 67 level matches as well.
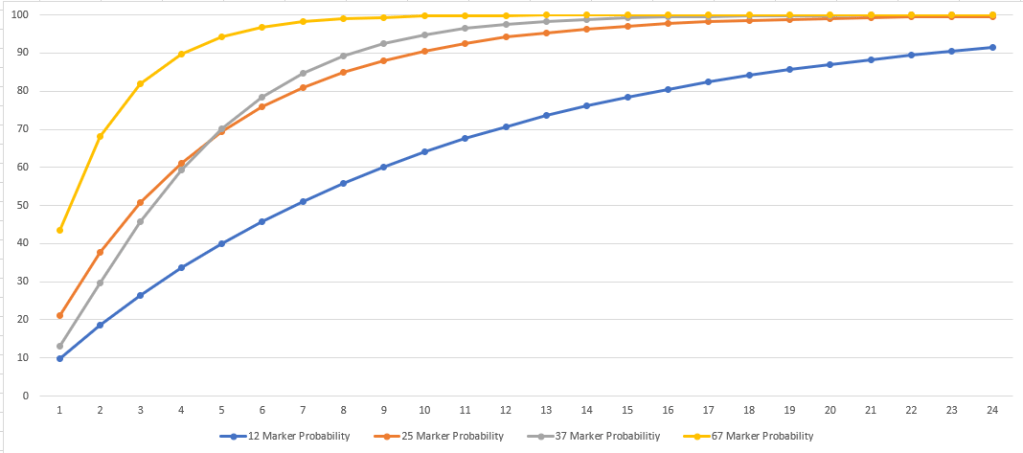
I find it interesting that the numbers suggest that a 37/37 match has a lower probability of having an MRCA within 1-4 generations than a 25/25 match does. I double checked the number the TiP Calculator was giving me to be sure this wasn’t some error I introduced.
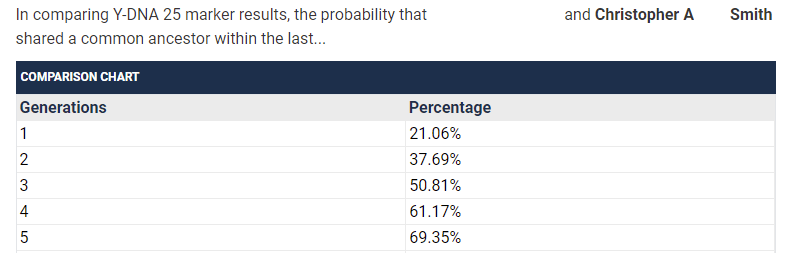
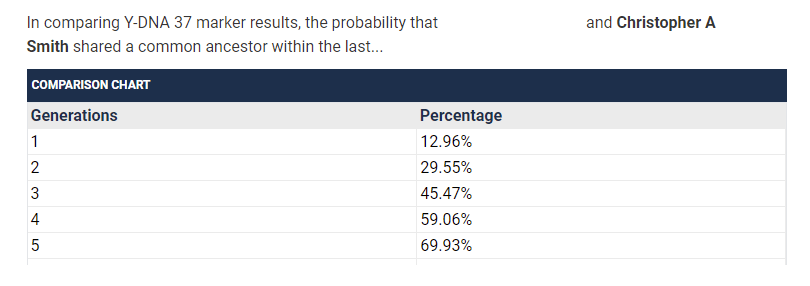
There is a field that can be adjusted if it is known that you and a match do not share a common ancestor in a number of generations. However, I’m inferring from the above FAQ note that this shouldn’t be done for yDNA matches who are of a genetic distance of 0?
These numbers begin to look very different.
But this is only FTDNA’s take on TMRCA estimates based on yDNA matching, limited to 24 generations. Let’s look at another source:
In looking at the above page, we find the following graph and table:
Method: B. Walsh, 2001. Estimating the time to the MRCA for the Y chromosome or mtDNA for a pair of individuals, Genetics 158: 897–912
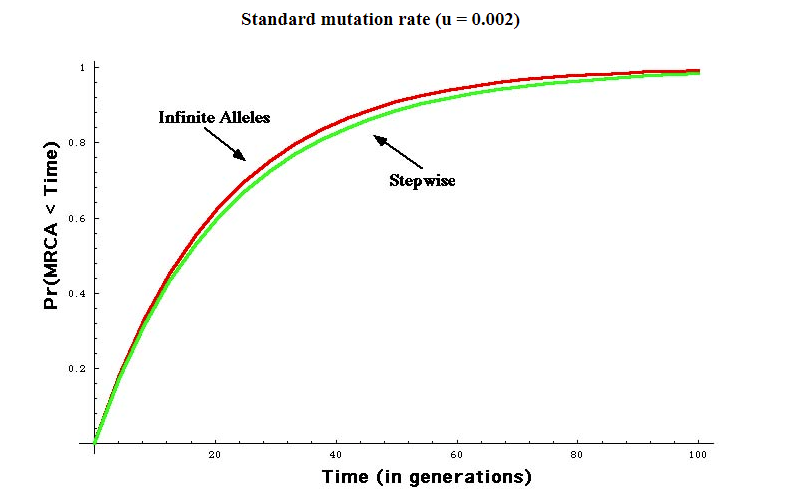
Both methods using Standard Mutation Rate:
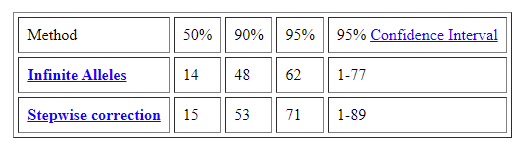
So this suggests that we enter the 90% confidence interval with a standard (slower?) mutation rate at around 48-53 generations, depending on which calculation model is used.
48 generations is, assuming around 25 years per generation, about 1200 years ago.
53 generations is, assuming around 25 years per generation, about 1325 years ago.
For folks with deep ancestry in England and that neighborhood, this could place, with 90% confidence, their TMRCA with a 12/12 yDNA match at around 300 years before the Norman Conquest, a time well before surnames were in use.
However, I want to show how even these estimates can be misleading.
One of my 63/67 yDNA matches, who has a close cousin who is a 67/67 yDNA match to myself, has done Y Haplogroup and BigY testing at FTDNA. They are shown to be R-DF41. I did an autosomal DNA test with 23andMe and they report my yDNA haplogroup as R-CTS2501. These values are synonymous.
According to yFull.com, R-DF41 aka R-CTS2501 emerged around 4100 years before present (ybp).
What this means is that, if only comparing yDNA haplogroups, if two men match, it can be said they have a TMRCA of around 4100 years ago, using R-DF41 as an example. While not super useful in and of itself, haplogroup matching can be useful for identifying potential yDNA testing candidates. People who autosomally match, are of the same yDNA haplogroup, and also share a surname or a common paper trail narrative make good candidates for further, more precise, 67 level STR testing and comparison.
R-CTS2501 is a subgroup of R-DF13, R-L21, R-S461, R-P312, R-L151, R-PF6538, R-L52, R-L51, R-L23, R-M269
I see one of my 12/12 matches is listed on FTDNA as R-L2. Let’s see where that fits in.
R-L2 is a subgroup of R-U152, R-P312, R-L151, R-PF6538, R-L52, R-L51, R-L23, R-M269
So based on this information that we are both part of R-P312, we can see that our TMRCA is actually waaay back at around 4,800 ybp. Assuming 25 years per generation, is it around 192 generations.
It appears that at least 4 of my 12/12 yDNA matches who are not Smiths are linked to R-P312. The remainder are generically R-M269.
I have also uploaded autosomal DNA kits for 67/67 yDNA matches who have not done yDNA haplogroup testing (and who also do not share any significant amount of autosomal DNA with myself) to the Morely yDNA Haplogroup/Subclade Predictor and found their results to match exactly the above information from FTDNA and 23andMe.
Ok, that’s all for now.
Chris
